Preparation and exploration of external control datasets
2021-10-01
Source:vignettes/01-data-prep.Rmd
01-data-prep.RmdSetup
The purpose of this analysis is to explore the data, specifically the (i) amount of missingness in potential covariates, (ii) small sample sizes among categories of categorical variables, and (iii) the distribution of potential covariates across the treated and control groups prior to any adjustment. The following R packages are used.
Data
The underlying dataset used for the analysis is built using the ecdata package. It consists of patients with advanced non-small cell lung cancer (NSCLC) and is documented here. The pins_nsclc() pins the datasets locally using pins::pin() so that the dataset only needs to be built once. We preprocess the data into a format more suitable for propensity score modeling with the preprocess() function and filter to remove the comparator arm from the RCT in order to emulate a single arm trial.
ecdata::pin_nsclc(update = FALSE) # Pins dataset locally
nsclc <- pin_get("nsclc") %>%
preprocess()
ec_rct1 <- nsclc %>% # Hypothetical single arm trial with external control
filter(!(source_type == "RCT" & arm_type == "Comparator"))All pairwise comparisons that will be used in the analyses are displayed in the table below.
analysis <- new_analysis(nsclc)
analysis %>%
html_table() %>%
scroll_box(width = "100%")| Number | ID | External | Trial | External | Trial | External control | Internal control | Experimental arm |
|---|---|---|---|---|---|---|---|---|
| 1 | nct02008227_fi_1 | Flatiron | NCT02008227 | Docetaxel | Atezolizumab | 526 | 425 | 425 |
| 2 | nct01903993_fi_1 | Flatiron | NCT01903993 | Docetaxel | Atezolizumab | 476 | 143 | 144 |
| 3 | nct02366143_fi_1 | Flatiron | NCT02366143 | Bevacizumab + Carboplatin | Atezolizumab + Bevacizumab + Carboplatin | 602 | 336 | 356 |
| 4 | nct01351415_fi_1 | Flatiron | NCT01351415 | SOC | Bevacizumab + SOC | 381 | 240 | 245 |
| 5 | nct01519804_fi_1 | Flatiron | NCT01519804 | Placebo + Platinum + Paclitaxel | MetMAb + Platinum + Paclitaxel | 1945 | 54 | 55 |
| 6 | nct01496742_fi_1 | Flatiron | NCT01496742 | Placebo + Bevacizumab + Platinum | MetMAb + Bevacizumab + Platinum + Paclitaxel | 956 | 70 | 69 |
| 7 | nct01496742_fi_2 | Flatiron | NCT01496742 | Placebo + Platinum + Pemetrexed | MetMAb + Platinum + Pemetrexed | 3244 | 61 | 59 |
| 8 | nct01366131_fi_1 | Flatiron | NCT01366131 | Placebo + Bevacizumab + Carboplatin + Paclitaxel | MEGF0444A + Bevacizumab + Carboplatin + Paclitaxel | 963 | 52 | 52 |
| 9 | nct01493843_fi_1 | Flatiron | NCT01493843 | Placebo (340 mg) + Carboplatin + Paclitaxel | Pictilisib (340 mg) + Carboplatin + Paclitaxel | 1274 | 125 | 126 |
| 10 | nct01493843_fi_2 | Flatiron | NCT01493843 | Placebo (340 mg) + Carboplatin + Paclitaxel + Bevacizumab | Pictilisib (340 mg) + Carboplatin + Paclitaxel + Bevacizumab | 953 | 79 | 79 |
| 11 | nct01493843_fi_3 | Flatiron | NCT01493843 | Placebo (260 mg) + Carboplatin + Paclitaxel + Bevacizumab | Pictilisib (260 mg) + Carboplatin + Paclitaxel + Bevacizumab | 953 | 30 | 62 |
| 12 | nct02367781_fi_1 | Flatiron | NCT02367781 | Nab-Paclitaxel + Carboplatin | Atezolizumab + Nab-Paclitaxel + Carboplatin | 87 | 228 | 451 |
| 13 | nct02367794_fi_1 | Flatiron | NCT02367794 | Nab-Paclitaxel + Carboplatin | Atezolizumab + Nab-Paclitaxel + Carboplatin | 497 | 340 | 343 |
| 14 | nct02657434_fi_1 | Flatiron | NCT02657434 | Carboplatin or Cisplatin + Pemetrexed | Atezolizumab + Carboplatin or Cisplatin + Pemetrexed | 1536 | 286 | 292 |
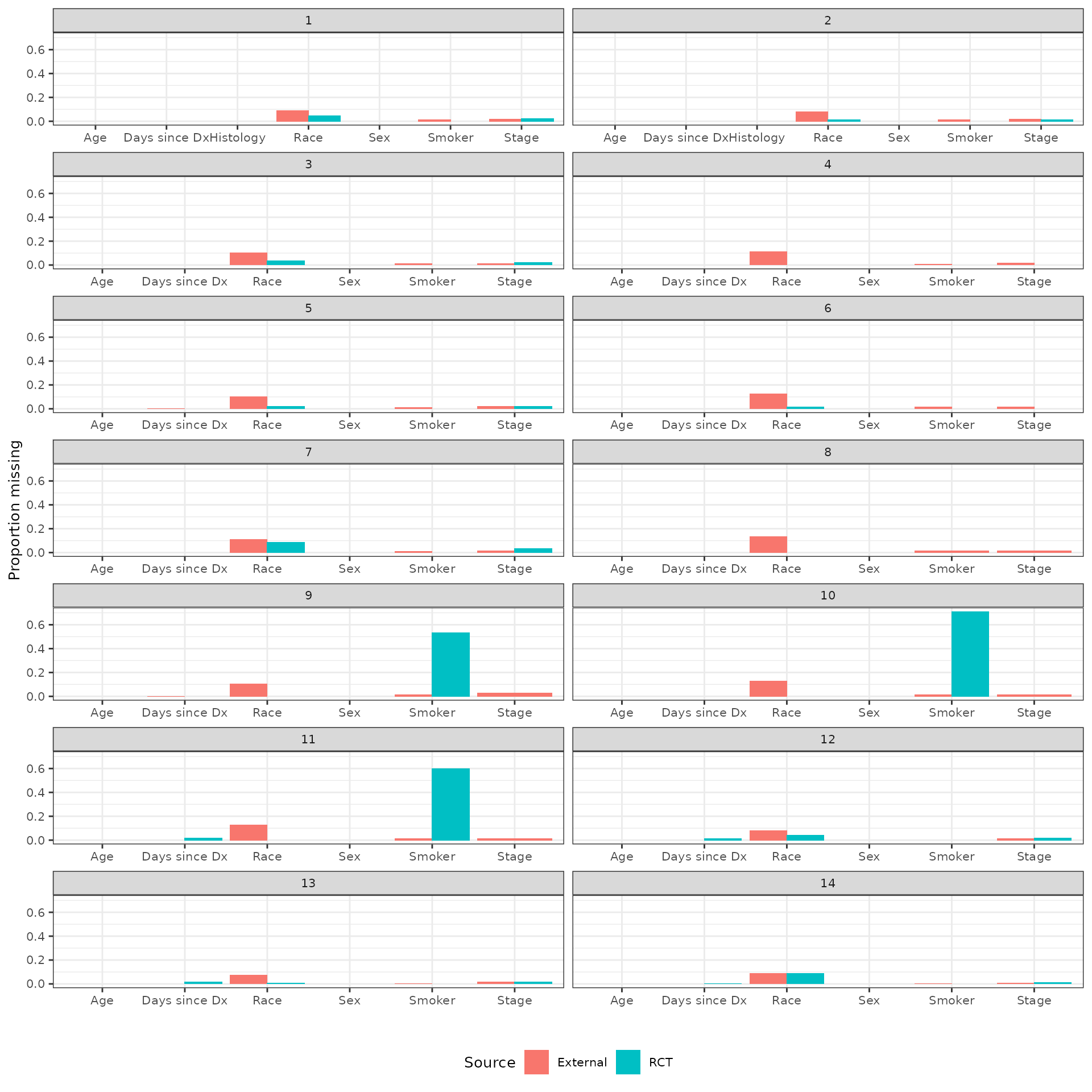
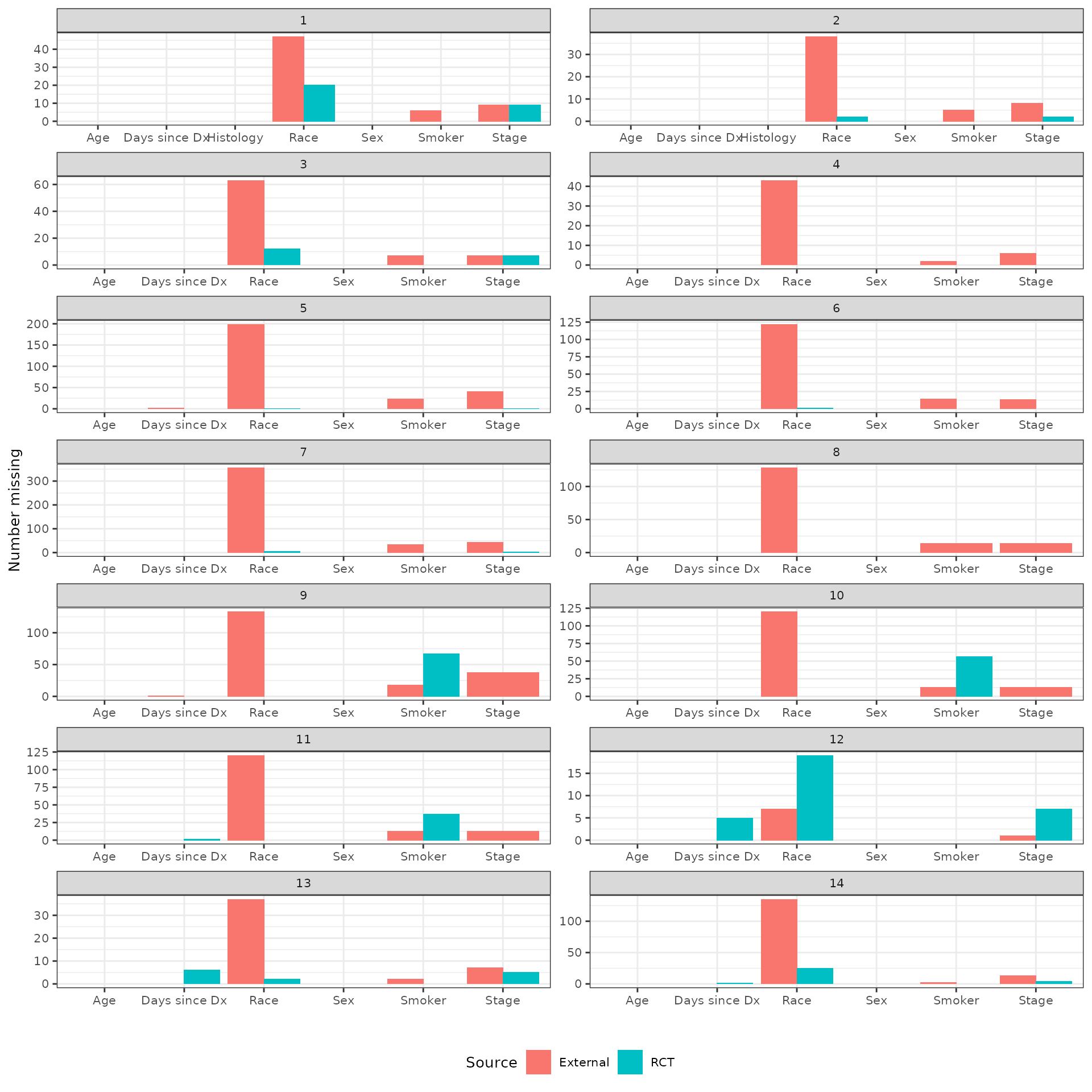
Missing data
We count the number of missing values of each potential covariate for each pairwise analysis. Both the number of missing and the proportion missing (of the total number of observations with each arm) are plotted.
n_missing_df <- count_missing(ec_rct1, vars = get_ps_vars())Distribution of covariates
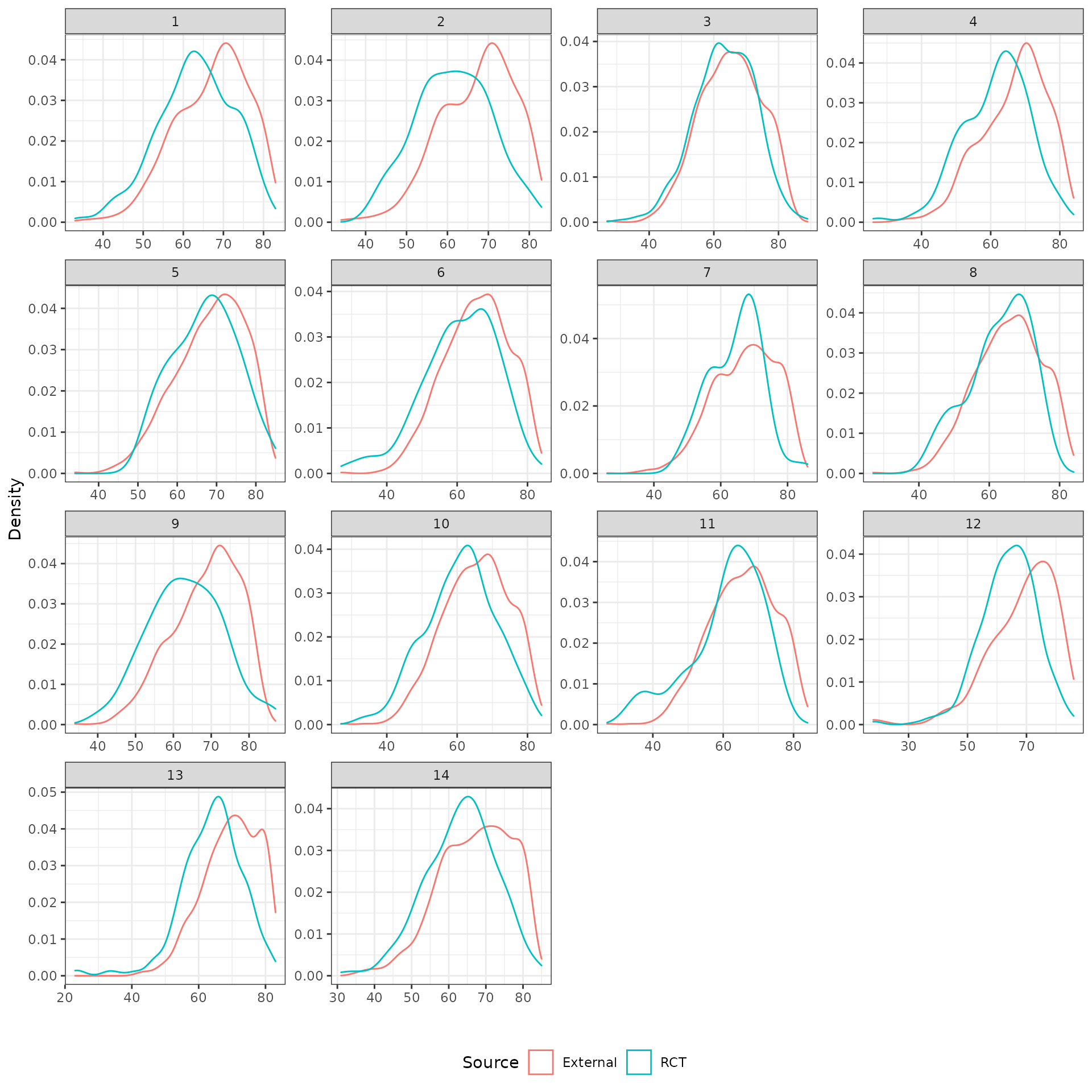
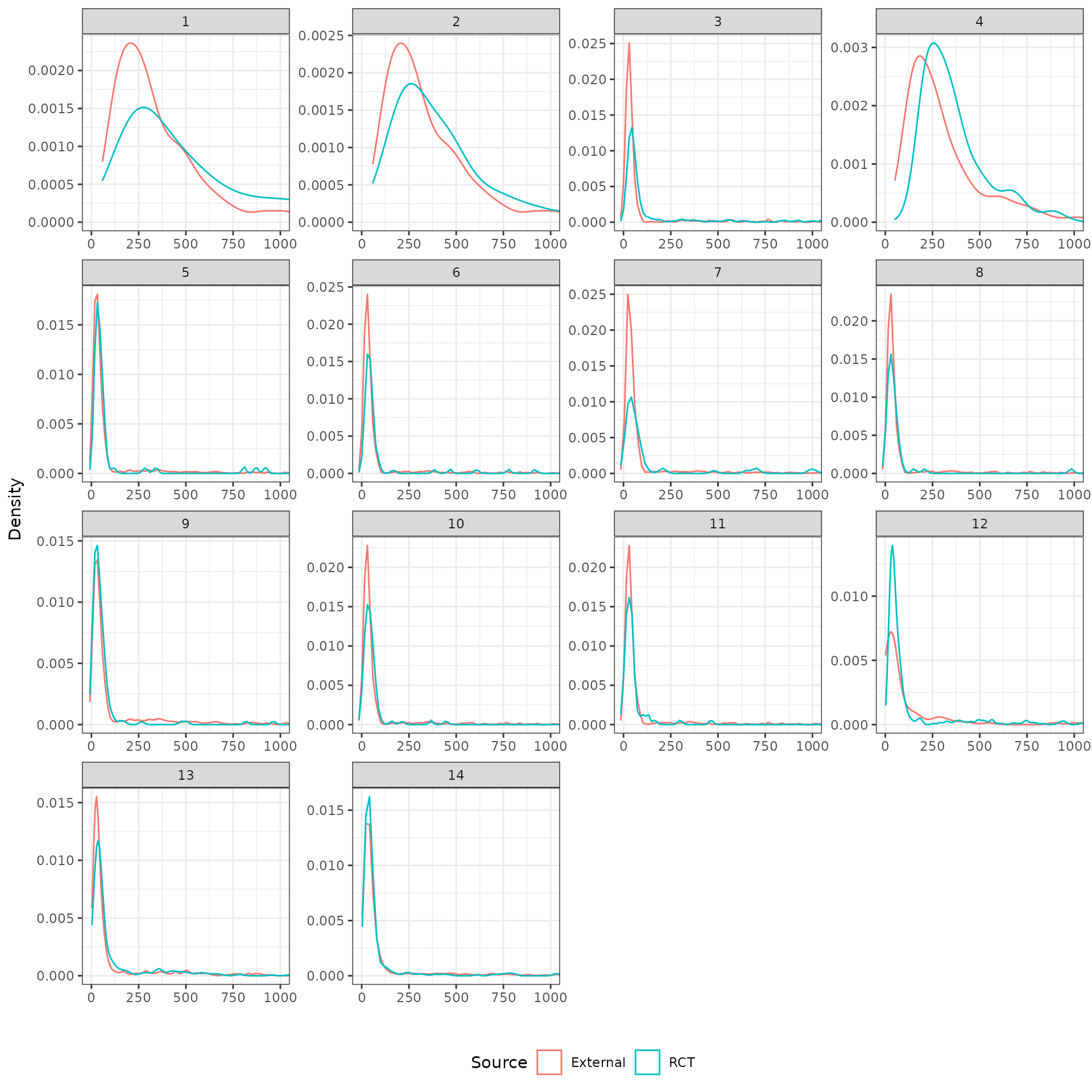
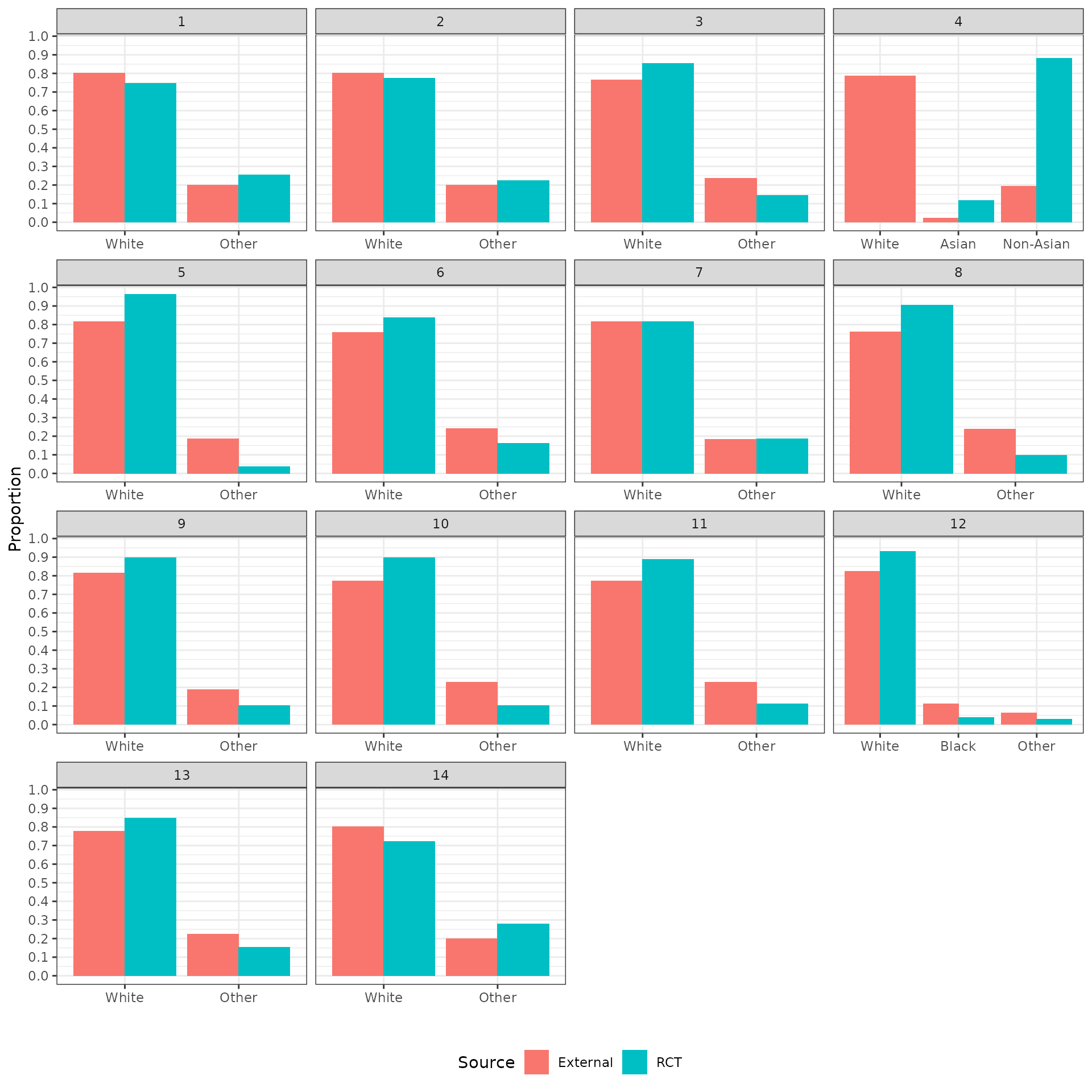
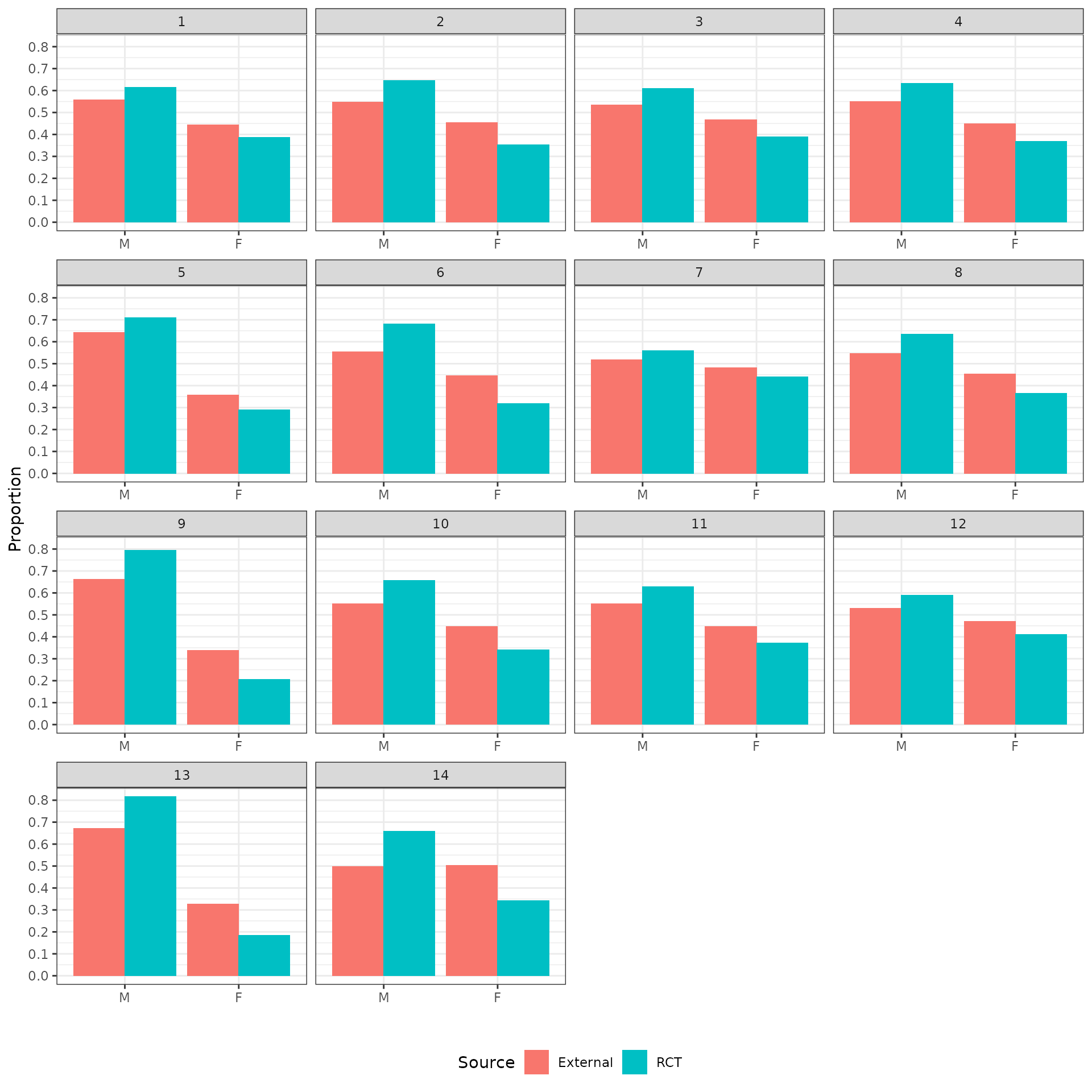
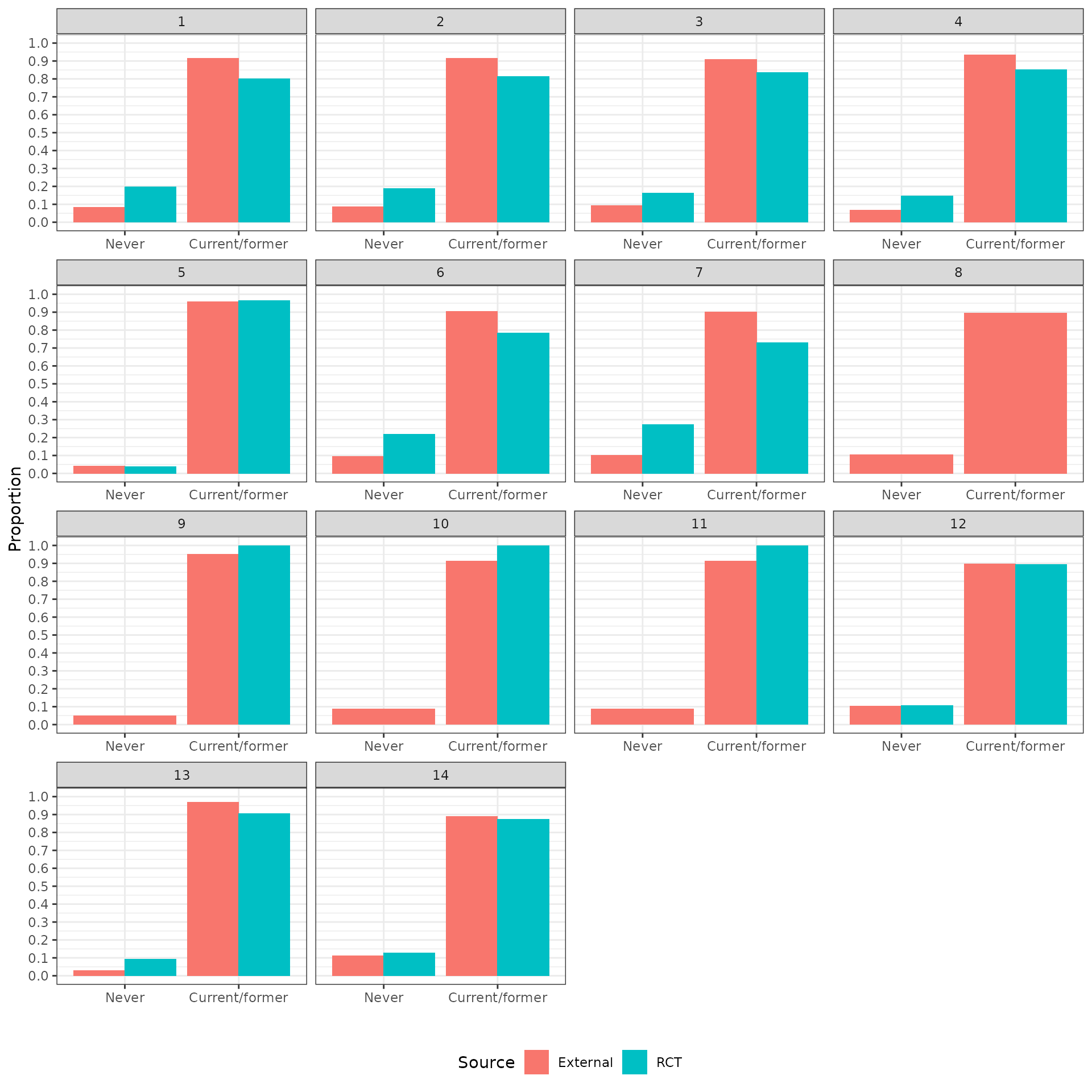
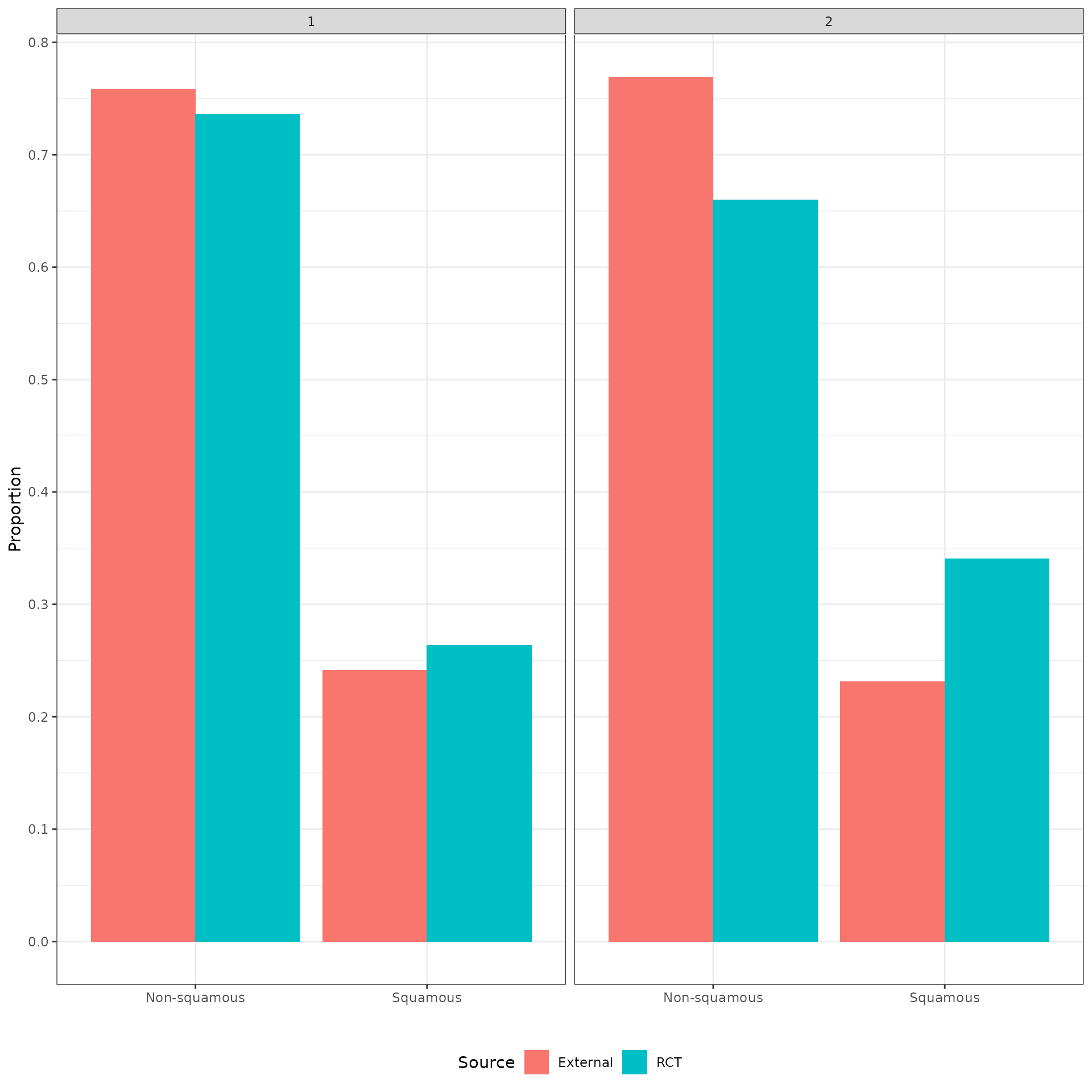
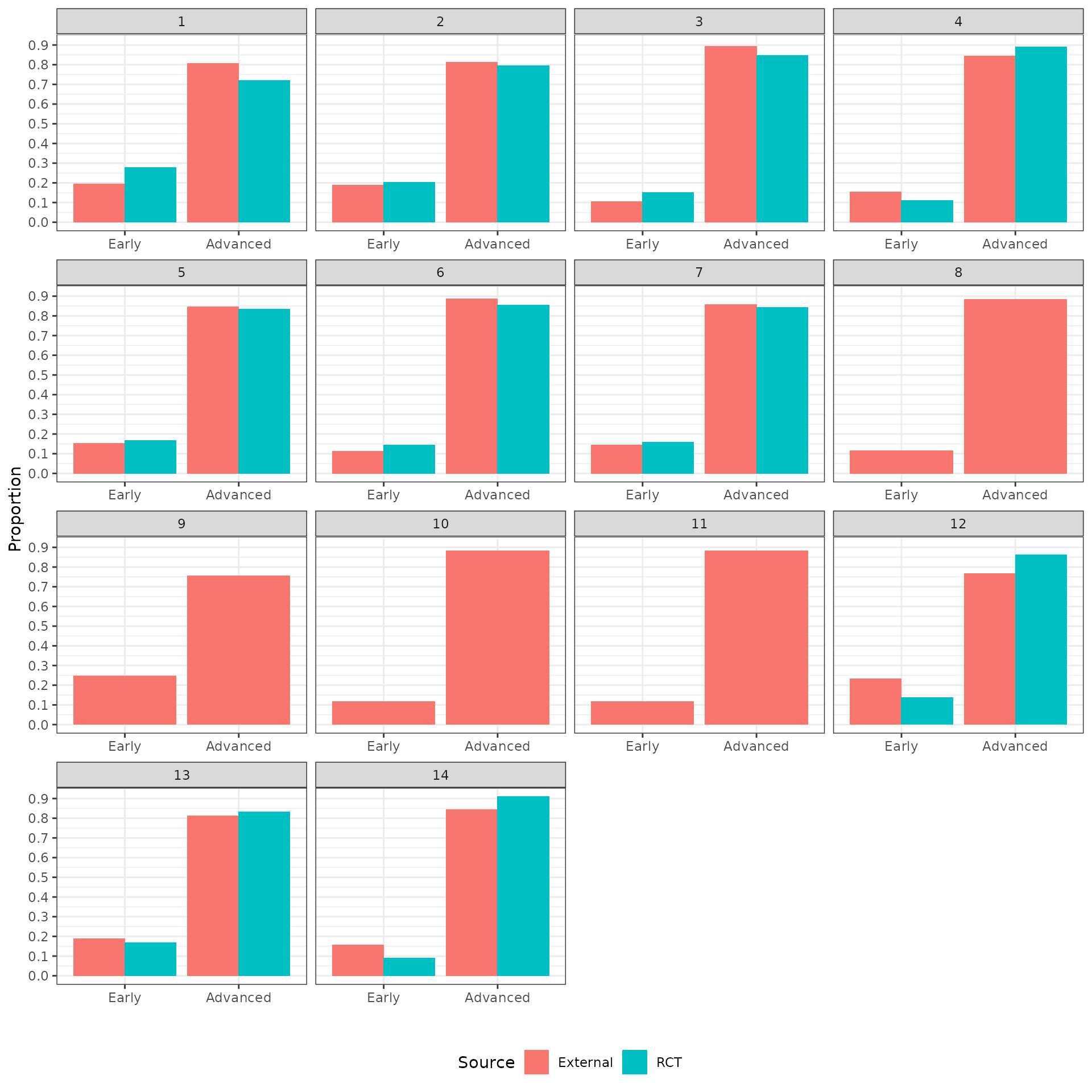
The distributions of the covariates in the unadjusted sample are plotted to check the degree of imbalance prior to adjustment and check for outliers or coding errors. There are a couple of points worth noting:
NCT01366131 did not collect smoking information.
It is not possible to determine whether a patient is a “Never” smoker in NCT01493843 because there is only a “Former/never” category. Smoking status can therefore not be used for propensity score analyses for this trial.
Patients in a race category with fewer than 10 observations were coded as “Other” race.
Categorical variables with small sample sizes
The plots above suggest that some categories of the categorical variables might still have small sample sizes even after preprocessing. Categories of categorical variables with fewer than 10 observations are shown in the table below.
count_by(ec_rct1, vars = get_ps_catvars(),
by = c("analysis_num", "source_type"),
max_n = 9) %>%
html_table()| analysis_num | source_type | var | value | n |
|---|---|---|---|---|
| 4 | External | race_grouped | Asian | 7 |
| 5 | RCT | race_grouped | Other | 2 |
| 5 | RCT | smoker_grouped | Never | 2 |
| 5 | RCT | stage_grouped | Early | 9 |
| 7 | RCT | stage_grouped | Early | 9 |
| 8 | RCT | race_grouped | Other | 5 |
| 10 | RCT | race_grouped | Other | 8 |
| 11 | RCT | race_grouped | Other | 7 |
| 12 | External | race_grouped | Black | 9 |
| 12 | External | race_grouped | Other | 5 |
| 12 | External | smoker_grouped | Never | 9 |
Save output
saveRDS(analysis, file = "analysis-data-prep.rds")Session information
## R version 4.0.0 (2020-04-24)
## Platform: x86_64-pc-linux-gnu (64-bit)
## Running under: Ubuntu 18.04.5 LTS
##
## Matrix products: default
## BLAS: /usr/lib/x86_64-linux-gnu/openblas/libblas.so.3
## LAPACK: /usr/lib/x86_64-linux-gnu/libopenblasp-r0.2.20.so
##
## locale:
## [1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
## [3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
## [5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
## [7] LC_PAPER=en_US.UTF-8 LC_NAME=C
## [9] LC_ADDRESS=C LC_TELEPHONE=C
## [11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
##
## attached base packages:
## [1] stats graphics grDevices utils datasets methods base
##
## other attached packages:
## [1] pins_0.4.5 kableExtra_1.3.4 ggplot2_3.3.5 ecmeta.nsclc_0.1.0
## [5] ecdata_0.2.1 dplyr_1.0.7
##
## loaded via a namespace (and not attached):
## [1] svglite_2.0.0 lattice_0.20-45 tidyr_1.1.4 assertthat_0.2.1
## [5] rprojroot_2.0.2 digest_0.6.28 utf8_1.2.2 R6_2.5.1
## [9] backports_1.2.1 evaluate_0.14 highr_0.9 httr_1.4.2
## [13] pillar_1.6.3 rlang_0.4.11 rstudioapi_0.13 data.table_1.14.2
## [17] jquerylib_0.1.4 Matrix_1.3-4 rmarkdown_2.11 pkgdown_1.6.1
## [21] labeling_0.4.2 textshaping_0.3.5 desc_1.4.0 splines_4.0.0
## [25] webshot_0.5.2 stringr_1.4.0 munsell_0.5.0 compiler_4.0.0
## [29] xfun_0.26 pkgconfig_2.0.3 systemfonts_1.0.2 htmltools_0.5.2
## [33] tidyselect_1.1.1 tibble_3.1.5 fansi_0.5.0 viridisLite_0.4.0
## [37] crayon_1.4.1 withr_2.4.2 rappdirs_0.3.3 grid_4.0.0
## [41] xtable_1.8-4 jsonlite_1.7.2 gtable_0.3.0 lifecycle_1.0.1
## [45] DBI_1.1.1 magrittr_2.0.1 scales_1.1.1 stringi_1.7.4
## [49] cachem_1.0.6 farver_2.1.0 fs_1.5.0 xml2_1.3.2
## [53] bslib_0.3.0 ellipsis_0.3.2 filelock_1.0.2 ragg_1.1.3
## [57] generics_0.1.0 vctrs_0.3.8 tools_4.0.0 glue_1.4.2
## [61] purrr_0.3.4 fastmap_1.1.0 survival_3.2-13 yaml_2.2.1
## [65] colorspace_2.0-2 rvest_1.0.1 memoise_2.0.0 knitr_1.36
## [69] sass_0.4.0